Introductory Morphology
“Moment is a word which most often refers to an ambiguously short length of time, but also signifies in mathematics a quantitative measure of the shape of a set of points, and in physics relates to the perpendicular distance from a point to a line or a surface.”
Wikiquote
import numpy as np
from matplotlib import pyplot as plt, cm
%matplotlib inline
import skdemo
plt.rcParams['image.cmap'] = 'cubehelix'
plt.rcParams['image.interpolation'] = 'none'
image = np.array([[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0]], dtype=np.uint8)
plt.imshow(image);

from skimage import morphology
sq = morphology.square(width=3)
dia = morphology.diamond(radius=1)
disk = morphology.disk(radius=2)
print(sq)
print('\n')
print(dia)
print('\n')
print(disk)
[[1 1 1]
[1 1 1]
[1 1 1]]
[[0 1 0]
[1 1 1]
[0 1 0]]
[[0 0 1 0 0]
[0 1 1 1 0]
[1 1 1 1 1]
[0 1 1 1 0]
[0 0 1 0 0]]
skdemo.imshow_all(image, morphology.erosion(image, sq), shape=(1, 2))

skdemo.imshow_all(image, morphology.dilation(image, sq))
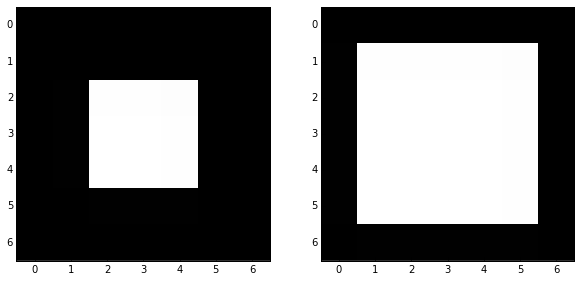
skdemo.imshow_all(image, morphology.erosion(image, dia))

skdemo.imshow_all(image, morphology.dilation(image, dia))
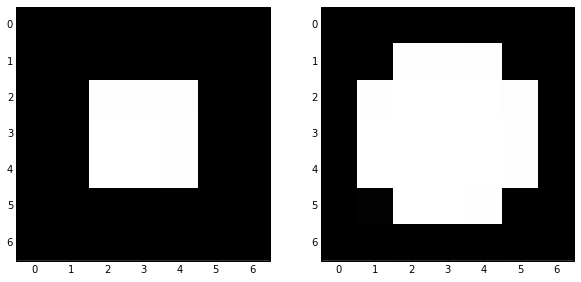
skdemo.imshow_all(image, morphology.erosion(image, disk))

skdemo.imshow_all(image, morphology.dilation(image, disk))
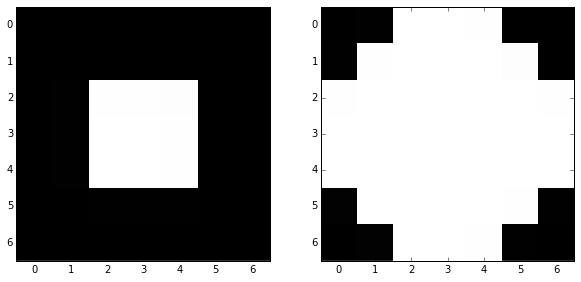
image = np.array([[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 1, 1, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], np.uint8)
plt.imshow(image);

What happens when you run an erosion followed by a dilation of this image?
What about the reverse?
Try to imagine the operations in your head before trying them out below.
skdemo.imshow_all(image, morphology.opening(image, sq)) # erosion -> dilation
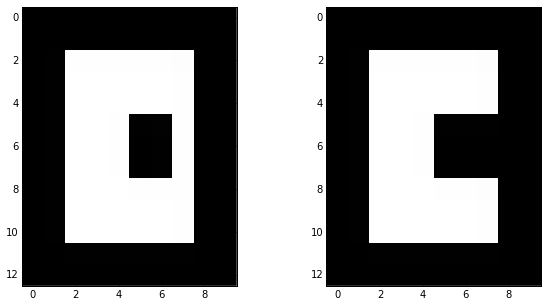
Opening result
skdemo.imshow_all(image, morphology.closing(image, sq)) # dilation -> erosion
Closing result
from skimage import data, color
hub = color.rgb2gray(data.hubble_deep_field()[350:450, 90:190])
plt.imshow(hub);

Remove the smaller objects to retrieve the large galaxy.
disk = morphology.disk(radius=2)
dst = morphology.erosion(hub, disk)
skdemo.imshow_with_histogram(dst)
(<matplotlib.axes._subplots.AxesSubplot at 0xadbe656c>,
<matplotlib.axes._subplots.AxesSubplot at 0xadf49f2c>)

blob = dst > 0.5
plt.imshow(blob);
<matplotlib.image.AxesImage at 0xadf3fe2c>

from scipy import ndimage
label_im, nb_labels = ndimage.label(blob)
How many regions?
nb_labels
1
plt.imshow(label_im);

Size
sizes = ndimage.sum(blob, label_im, range(nb_labels+1))
print('sizes = ', sizes)
sizes = [ 0. 605.]
Center of Mass
print(ndimage.center_of_mass(blob))
(51.292079207920793, 33.214521452145213)
h,k = ndimage.center_of_mass(blob)
blob[h,k] = 0
plt.imshow(blob);

Written on April 5, 2015