SCIKIT's Image Segmentation
“Research is to see what everybody else has seen, and to think what nobody else has thought.”
Albert Szent-Gyorgyi
import numpy as np
%matplotlib inline
from matplotlib import pyplot as plt
import skdemo
plt.rcParams['image.cmap'] = 'spectral'
from skimage import io, segmentation as seg, color
url = '../images/spice_1.jpg'
image = io.imread(url)
labels = seg.slic(image, n_segments=18, compactness=10)
skdemo.imshow_all(image, labels.astype(float) / labels.max())
print(labels)
[[ 0 0 0 ..., 4 4 4]
[ 0 0 0 ..., 4 4 4]
[ 0 0 0 ..., 4 4 4]
...,
[15 15 15 ..., 19 19 19]
[15 15 15 ..., 19 19 19]
[15 15 15 ..., 19 19 19]]

labels = seg.slic(image, n_segments=18, compactness=40)
skdemo.imshow_all(image, labels.astype(float) / labels.max())
print(labels)
[[ 0 0 0 ..., 4 4 4]
[ 0 0 0 ..., 4 4 4]
[ 0 0 0 ..., 4 4 4]
...,
[15 15 15 ..., 19 19 19]
[15 15 15 ..., 19 19 19]
[15 15 15 ..., 19 19 19]]

labels = seg.slic(image, n_segments=18, compactness=100)
skdemo.imshow_all(image, labels.astype(float) / labels.max())
print(labels)
[[ 0 0 0 ..., 4 4 4]
[ 0 0 0 ..., 4 4 4]
[ 0 0 0 ..., 4 4 4]
...,
[15 15 15 ..., 19 19 19]
[15 15 15 ..., 19 19 19]
[15 15 15 ..., 19 19 19]]
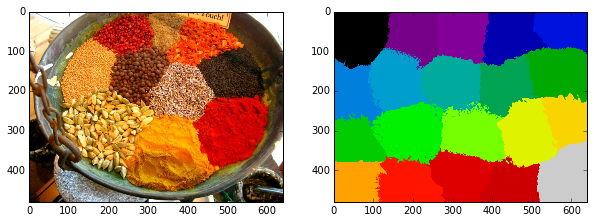
def mean_color(image, labels):
out = np.zeros_like(image)
for label in np.unique(labels):
indices = np.nonzero(labels == label)
out[indices] = np.mean(image[indices], axis=0)
return out
skdemo.imshow_all(image, mean_color(image, labels))

labels = seg.slic(image, n_segments=24, compactness=40,
sigma=2, enforce_connectivity=True)
label_image = mean_color(image, labels)
skdemo.imshow_all(image, label_image)

def myfunc(*args, **kwargs):
labels = seg.slic(image, *args, **kwargs)
color = mean_color(image, labels)
skdemo.imshow_all(image, color)
from IPython.html import widgets
w = widgets.interactive(myfunc, compactness = (5, 200, 10), n_segments =(25, 200, 15), enforce_connectivity = True)
w.msg_throttle = 1
w

def myfunc(*args, **kwargs):
labels = seg.slic(image, *args, **kwargs)
color = mean_color(image, labels)
skdemo.imshow_all(image, color)
from IPython.html import widgets
w = widgets.interactive(myfunc, compactness = (5, 100, 5), n_segments =(25, 200, 15), sigma = (0.,5.,0.1), enforce_connectivity = True)
w.msg_throttle = 1
w

Image types: boundary images
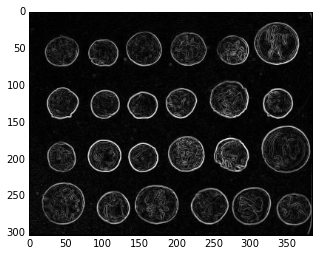
Often, the contrast between regions is not sufficient to distinguish them, but there is a clear boundary between the two. Using an edge detector on these images, followed by a watershed, often gives very good segmentation.
from skimage import data, filters as filters
from matplotlib import pyplot as plt, cm
coins = data.coins()
edges = filters.sobel(coins)
plt.imshow(edges, cmap='gray');
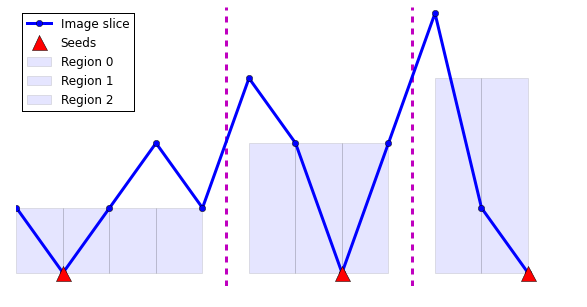
The watershed algorithm finds the regions between these edges. It does so by envisioning the pixel intensity as height on a topographic map. It then “floods” the map from the bottom up, starting from seed points. These flood areas are called “watershed basins” and when they meet, they form the image segmentation.
from skimage.morphology import watershed
from scipy import ndimage as nd
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11])
y = np.array([1, 0, 1, 2, 1, 3, 2, 0, 2, 4, 1, 0])
seeds = nd.label(y == 0)[0]
seed_positions = np.argwhere(seeds)[:, 0]
print("Seeds:", seeds)
print("Seed positions:", seed_positions)
# ------------------------------- #
result = watershed(y, seeds)
# ------------------------------- #
# You can ignore the code below--it's just
# to make a pretty plot of the results.
plt.figure(figsize=(10, 5))
plt.plot(y, '-o', label='Image slice', linewidth=3)
plt.plot(seed_positions, np.zeros_like(seed_positions), 'r^',
label='Seeds', markersize=15)
for n, label in enumerate(np.unique(result)):
mask = (result == label)
plt.bar(x[mask][:-1], result[mask][:-1],
width=1, label='Region %d' % n,
alpha=0.1)
plt.vlines(np.argwhere(np.diff(result)) + 0.5, -0.2, 4.1, 'm',
linewidth=3, linestyle='--')
plt.legend(loc='upper left', numpoints=1)
plt.axis('off')
plt.ylim(-0.2, 4.1);
Seeds: [0 1 0 0 0 0 0 2 0 0 0 3]
Seed positions: [ 1 7 11]
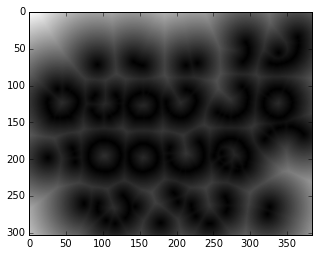
from scipy import ndimage as nd
threshold = 0.4
# Euclidean distance transform
# How far do we have to travel from a non-edge to find an edge?
non_edges = (edges < threshold)
distance_from_edge = nd.distance_transform_edt(non_edges)
plt.imshow(distance_from_edge, cmap='gray');
threshold = 0.35
# Euclidean distance transform
# How far do we have to travel from a non-edge to find an edge?
non_edges = (edges < threshold)
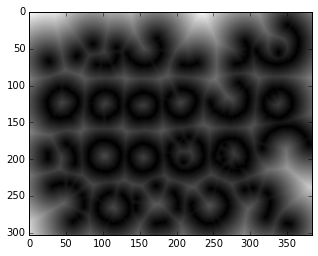
distance_from_edge = nd.distance_transform_edt(non_edges)
plt.imshow(distance_from_edge, cmap='gray');
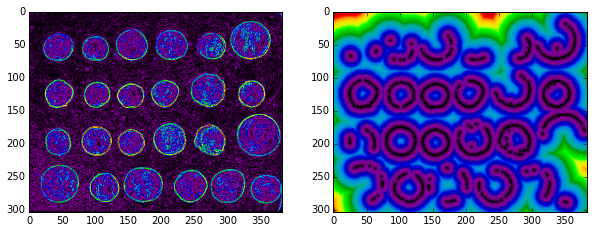
skdemo.imshow_all(edges/edges.max(), distance_from_edge/distance_from_edge.max());
from skimage import feature
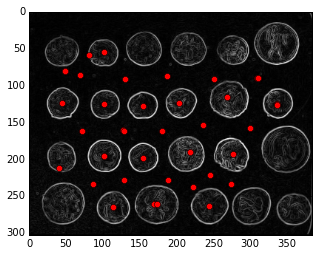
# -------------------------------------------------#
peaks = feature.peak_local_max(distance_from_edge)
print("Peaks shape:", peaks.shape)
# -------------------------------------------------#
peaks_image = np.zeros(coins.shape, np.bool)
peaks_image[tuple(np.transpose(peaks))] = True
seeds, num_seeds = nd.label(peaks_image)
plt.imshow(edges, cmap='gray')
plt.plot(peaks[:, 1], peaks[:, 0], 'ro');
plt.axis('image')
Peaks shape: (40, 2)
(-0.5, 383.5, 302.5, -0.5)
ws = watershed(edges, seeds)
from skimage import color
plt.imshow(color.label2rgb(ws, coins));
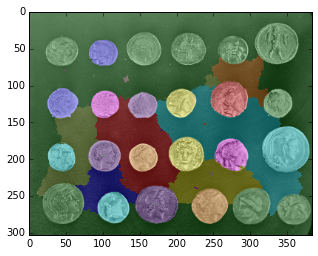
from skimage.measure import regionprops
regions = regionprops(ws)
ws_updated = ws.copy()
for region in regions:
if region.eccentricity > 0.6:
ws_updated[ws_updated == region.label] = 0
plt.imshow(color.label2rgb(ws_updated, coins, bg_label=0));

Written on April 6, 2015